ITK Release 4/Data Collection: Difference between revisions
From KitwarePublic
Jump to navigationJump to search
| (27 intermediate revisions by 6 users not shown) | |||
| Line 17: | Line 17: | ||
= Expected Data = | = Expected Data = | ||
* Maintain data using original acquisition format (Bill's suggestion)? | |||
{| border="1" | {| border="1" | ||
| Line 22: | Line 25: | ||
! Group !! Expected Amount of Data !! File Formats !! Features | ! Group !! Expected Amount of Data !! File Formats !! Features | ||
|- | |- | ||
| Raghu Machiraju || 10 Gb || TIFF || | | Raghu Machiraju || 10 Gb || TIFF, LSM, MetaImage || | ||
|- | |- | ||
| Ziv Yaniv || | | Ziv Yaniv || 10 Gb || DICOM, MetaImage (e.g. segmentation label maps), bmp/jpg/avi (2D frame grabbed US and x-ray fluoroscopy images)|| | ||
|- | |- | ||
| Brad Lowekamp || 10 Gb || TIFF ? || | | Brad Lowekamp || 10 Gb || TIFF ? || | ||
|- | |- | ||
| Hans Johnson || | | Hans Johnson || 4 Gb || GE45, Phillips, Siemens, Dicom, Analyze, Nifti, etc Migration from : http://www.nitrc.org/projects/brainstestdata/ || Dictionary of expected meta-information (spacing, size, data-type, etc) | ||
|- | |- | ||
| Sean Megason || | | Sean Megason || 10 Tb || TIFF, LSM [[File:Megason-A2D2proposal-SCORExx.pdf]] || | ||
|- | |||
| Marcel Pratwsaw || 10 Gb || ?? || | |||
|- | |||
| Marc Niethammer || 10 Gb || TIFF || Point spread functions (and image, or 12 (key,value) pair parameters | |||
|- | |- | ||
|} | |} | ||
= MetaData = | = MetaData = | ||
* | ==Medical== | ||
* physical quantities associated with image acquisition apparatus (e.g. camera calibration for endoscopy and x-ray, US calibration). | |||
* reason for scan (e.g. suspected liver tumor), should be in header but often is not. | |||
* contrast phase (e.g. pre contrast, arterial phase). | |||
* respiratory phase (e.g. end inspiration, end expiration). | |||
* apparatus used to acquire images when not specified in DICOM (e.g. xyz frame grabber). | |||
* rigid transformation data acquired with a tracker (pose of US/endoscope relative to tracking system, required for positioning multiple 2D images in 3D space) | |||
* calibration data of other equipment used in experiment (calibration of tracked pointer) | |||
= Structure = | = Structure = | ||
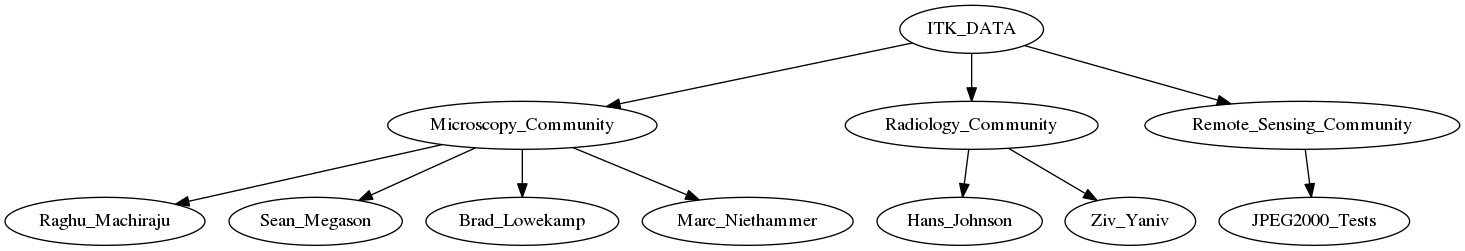
| Line 50: | Line 63: | ||
Microscopy_Community -> Sean_Megason | Microscopy_Community -> Sean_Megason | ||
Microscopy_Community -> Brad_Lowekamp | Microscopy_Community -> Brad_Lowekamp | ||
Microscopy_Community -> Marc_Niethammer | |||
ITK_DATA -> Radiology_Community | ITK_DATA -> Radiology_Community | ||
Radiology_Community -> Hans_Johnson | Radiology_Community -> Hans_Johnson | ||
Radiology_Community -> Ziv_Yaniv | Radiology_Community -> Ziv_Yaniv | ||
ITK_DATA -> Remote_Sensing_Community | |||
Remote_Sensing_Community -> JPEG2000_Tests | |||
} | } | ||
</graphviz> | </graphviz> | ||
== Strawman of data organization == | |||
Filename scheme | |||
AllSubjects/Subject/ScanDateTime/Modality/Subject_ScanDate_Modality.nii.gz | |||
ScanDate is in the form YYYYMMDDTT where we only keep 2 digits of time. | |||
so you would have e.g. | |||
ITKTestCTdata1/0001/2010122200/T1/0001_2010122200_T1.nii.gz | |||
= MIDAS Installations = | = MIDAS Installations = | ||
| Line 65: | Line 93: | ||
** http://midas.kitware.com/community/view/7 (ITK Testing Data) | ** http://midas.kitware.com/community/view/7 (ITK Testing Data) | ||
** http://midas.kitware.com/community/view/22 (Tutorials: Virtual Appliances > 2Gb) | ** http://midas.kitware.com/community/view/22 (Tutorials: Virtual Appliances > 2Gb) | ||
= Integration with CTest = | |||
* http://review.source.kitware.com/#change,301 | |||
* http://www.kitware.com/midaswiki/index.php/MIDAS%2BCTest | |||
== BrainWeb Datasets == | |||
* http://mouldy.bic.mni.mcgill.ca/brainweb/ | |||
* http://midas.kitware.com/collection/view/44 | |||
== XNAT == | |||
* http://central.xnat.org/app/template/Index.vm | |||
= Data Licenses = | |||
* http://www.kitware.com/blog/home/post/72 | |||
** Public Domain Dedication and License (PDDL) — “Public Domain for data/databases” http://www.opendatacommons.org/licenses/pddl/ | |||
** Attribution License (ODC-By) — “Attribution for data/databases” http://www.opendatacommons.org/licenses/by/ | |||
** Open Database License (ODC-ODbL) — “Attribution Share-Alike for data/databases” http://www.opendatacommons.org/licenses/odbl/ | |||
* Designed by the Scientific Commons at http://www.opendatacommons.org/ | |||
Latest revision as of 16:44, 20 December 2010
Overview
Testing Data
MIDAS Structure
- Communities (recursive)
- Collection
- Items
- Resources
- Bitstreams
- Bitstreams
- Resources
- Items
- Collection
Expected Data
- Maintain data using original acquisition format (Bill's suggestion)?
| Group | Expected Amount of Data | File Formats | Features |
|---|---|---|---|
| Raghu Machiraju | 10 Gb | TIFF, LSM, MetaImage | |
| Ziv Yaniv | 10 Gb | DICOM, MetaImage (e.g. segmentation label maps), bmp/jpg/avi (2D frame grabbed US and x-ray fluoroscopy images) | |
| Brad Lowekamp | 10 Gb | TIFF ? | |
| Hans Johnson | 4 Gb | GE45, Phillips, Siemens, Dicom, Analyze, Nifti, etc Migration from : http://www.nitrc.org/projects/brainstestdata/ | Dictionary of expected meta-information (spacing, size, data-type, etc) |
| Sean Megason | 10 Tb | TIFF, LSM File:Megason-A2D2proposal-SCORExx.pdf | |
| Marcel Pratwsaw | 10 Gb | ?? | |
| Marc Niethammer | 10 Gb | TIFF | Point spread functions (and image, or 12 (key,value) pair parameters |
MetaData
Medical
- physical quantities associated with image acquisition apparatus (e.g. camera calibration for endoscopy and x-ray, US calibration).
- reason for scan (e.g. suspected liver tumor), should be in header but often is not.
- contrast phase (e.g. pre contrast, arterial phase).
- respiratory phase (e.g. end inspiration, end expiration).
- apparatus used to acquire images when not specified in DICOM (e.g. xyz frame grabber).
- rigid transformation data acquired with a tracker (pose of US/endoscope relative to tracking system, required for positioning multiple 2D images in 3D space)
- calibration data of other equipment used in experiment (calibration of tracked pointer)
Structure
Suggested Organization
Strawman of data organization
Filename scheme
AllSubjects/Subject/ScanDateTime/Modality/Subject_ScanDate_Modality.nii.gz
ScanDate is in the form YYYYMMDDTT where we only keep 2 digits of time.
so you would have e.g.
ITKTestCTdata1/0001/2010122200/T1/0001_2010122200_T1.nii.gz
MIDAS Installations
- http://www.insight-journal.org/midas/
- http://www.insight-journal.org/midas/community/view/22 (Miscellaneous collection of ITK related data)
- http://www.insight-journal.org/rire/ (RIRE: Retrospective Registration Evaluation)
- http://midas.kitware.com/
- http://midas.kitware.com/community/view/7 (ITK Testing Data)
- http://midas.kitware.com/community/view/22 (Tutorials: Virtual Appliances > 2Gb)
Integration with CTest
- http://review.source.kitware.com/#change,301
- http://www.kitware.com/midaswiki/index.php/MIDAS%2BCTest
BrainWeb Datasets
XNAT
Data Licenses
- http://www.kitware.com/blog/home/post/72
- Public Domain Dedication and License (PDDL) — “Public Domain for data/databases” http://www.opendatacommons.org/licenses/pddl/
- Attribution License (ODC-By) — “Attribution for data/databases” http://www.opendatacommons.org/licenses/by/
- Open Database License (ODC-ODbL) — “Attribution Share-Alike for data/databases” http://www.opendatacommons.org/licenses/odbl/
- Designed by the Scientific Commons at http://www.opendatacommons.org/