TubeTK/Events/2010.10.19: Difference between revisions
From KitwarePublic
Jump to navigationJump to search
(Created page with "* Dashboard: https://www.kitware.com/CDash/index.php?project=TubeTK ** Registration re-write needed * Wiki Documentation: http://public.kitware.com/Wiki/TubeTK = Stephen = == Co...") |
Jamie.snape (talk | contribs) No edit summary |
||
| (7 intermediate revisions by 3 users not shown) | |||
| Line 18: | Line 18: | ||
= Romain = | = Romain = | ||
* Implementation on the ComputeTortuosity module (only Distance Metric) | |||
** Started debugging | |||
*Future | |||
** More debugging! ---> hopefully testing soon | |||
** #tube command | |||
** Implementation of the other 2 methods : ICM SOAM | |||
= Danielle = | = Danielle = | ||
== Sliding organ registration == | |||
* Fixed "duh" problem from Thursday - extracting tangential and normal components before computing regularization | |||
* Regularization smoothing in normal direction completed | |||
* Added intensity distance function (mean squares) | |||
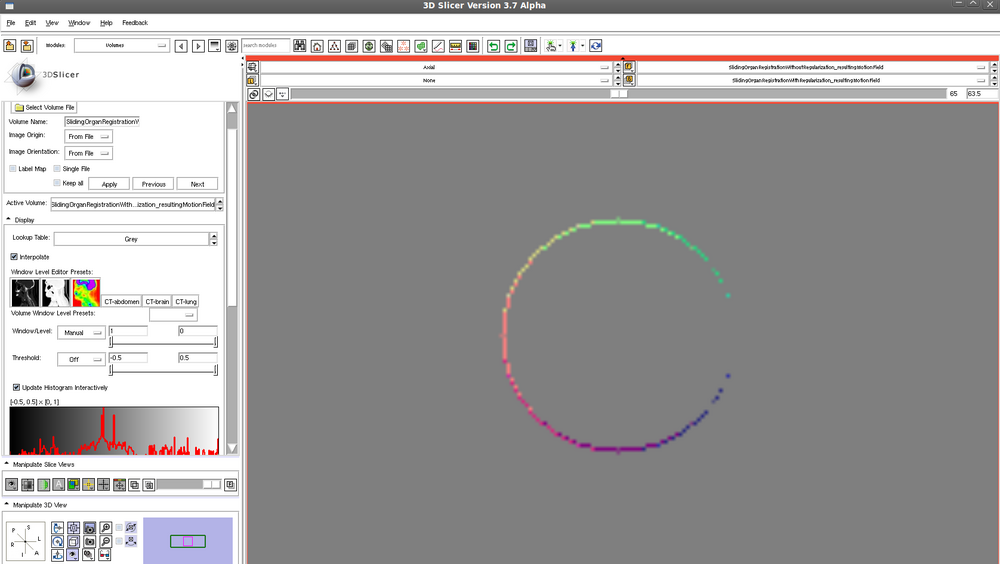
Translated sphere - motion field following registration WITHOUT regularization: | |||
[[File:translatedSphereMotionFieldWithoutRegularization.png| 1000px]] | |||
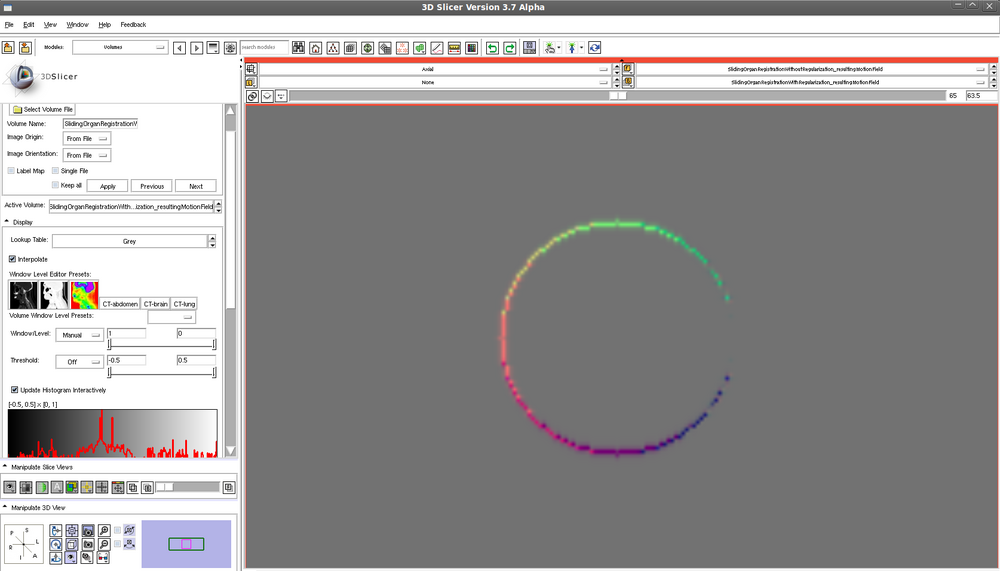
Translated sphere - motion field following registration WITH regularization: | |||
[[File:translatedSphereMotionFieldWithRegularization.png| 1000px]] | |||
== XCAT phantom == | |||
* Got XCAT running on my machine (thanks Andinet!) | |||
* Learned XCAT parameters | |||
* Wrote isbiValidation.py: | |||
** --basic (XCAT mode 0 - phantom mode): with respiratory period 5s and cardiac period 1s, generates two images at 0s and 2.5 s, saves as .mhd | |||
** --gold (XCAT mode 2 - spherical lesion generation mode): creates gold standard motion field, and saves as .txt (index, vector in 6 columns). Then run itkImageToImageDiffusiveDeformableRegistrationGenerateGoldStandardTest to create itk::Image<VectorType, Dimension> .mhd for visualization | |||
*** XCAT phantom gives gold standard vectors for surface borders (XCAT mode 4), but no internal vectors! So... | |||
*** for index in range - generate spherical lesion at index, use nurbs_save = 1 to save lesion.nrb, then calculate centroid of lesion control points | |||
[[File:2010_10_19_motionFieldParaviewOverview.png|800 px]] | |||
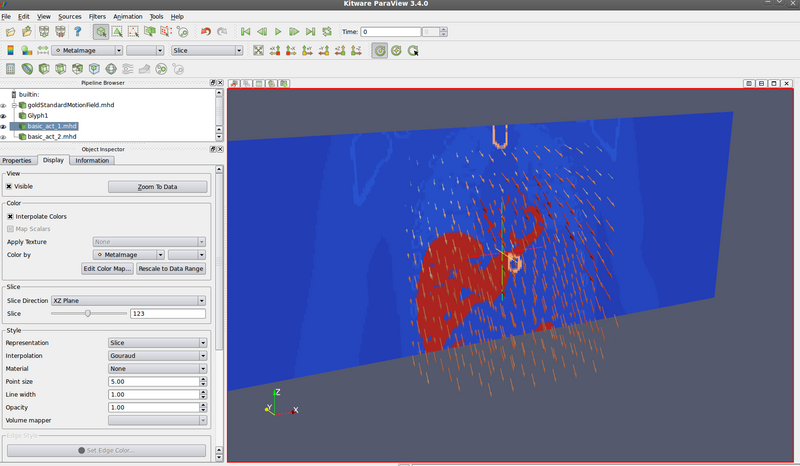
*** Question: are there discontinuities / sliding motion. Looks like it =) See circled region: | |||
[[File:2010_10_19_goldStandardMotionFieldDiscontinuities_annotated.png|800 px]] | |||
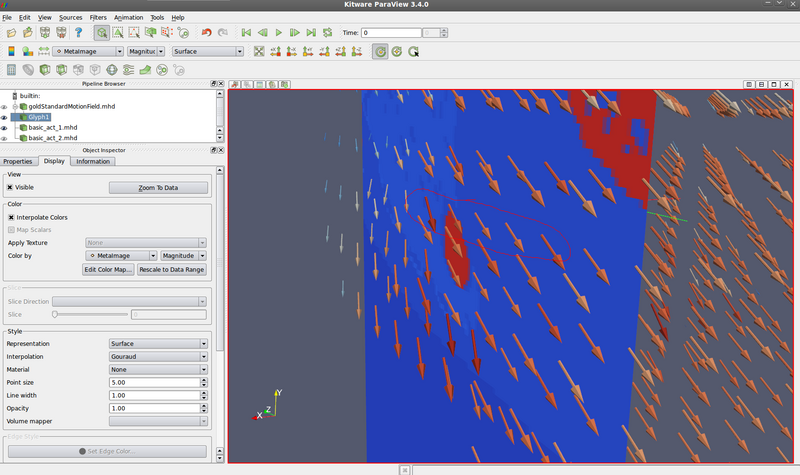
*** Question: does the motion field make sense? Yes: | |||
Fixed image (look at origin of vectors): | |||
[[File:2010_10_19_motionFieldParaview_1.png|800 px]] | |||
Moving image (look at destination of vectors): | |||
[[File:2010_10_19_motionFieldParaview_2.png|800 px]] | |||
** --segmentation (XCAT mode 5 - save anatomical variation mode, or run XCAT mode 0 with save_nurbs = 1) to save surfaces (.nrb text file). Then create new .txt file for the surface you want. Then run itkImageToImageDiffusiveDeformableRegistrationGenerateNormalsTest to create itk::Image<PixelType, Dimension> of control points | |||
*** Unfortunately: (1) control points need interpolation, can't find NURBS support in VTK; (2) XCAT has MANY types of surfaces for one organ - ex all intracardiac surfaces, etc, and it would be a lot of work to sort through them; (3) Perfect segmentation not great for registration method evaluation - removes segmentation error component of total error. | |||
*** Ex: right lung | |||
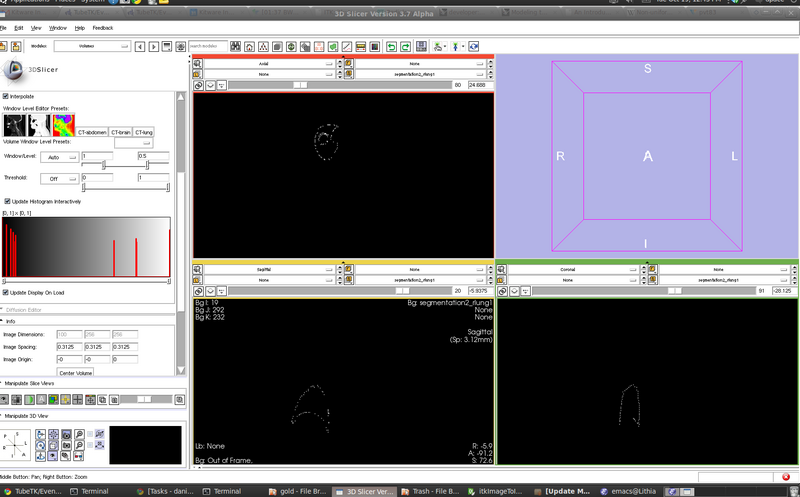
[[File:segmentation2_rlung.png | 800 px]] | |||
== Talking points == | |||
* Underlying XCAT deformation fields too perfect? They are likely interpolating the border vectors. | |||
== Future == | |||
* Boundary conditions | |||
* Compute weighting term = distance to organ border | |||
* Extract organ border normals from input data | |||
* Run registration + compare to gold standard for validation #s | |||
[[Category:TubeTK Events and Meetings|2010.10.19]] | |||
Latest revision as of 18:48, 26 July 2013
- Dashboard: https://www.kitware.com/CDash/index.php?project=TubeTK
- Registration re-write needed
- Wiki Documentation: http://public.kitware.com/Wiki/TubeTK
Stephen
Completed
- Ridge extractor success
- Email requests for TubeTK
- Licensing changes
- Blur3D -> Blur
Ongoing
- Radius extractor tests
- 2D and 3D only :(
- Funding re-allocation
Upcoming
- Application in plant biology
- Proposal with Paul / Liz
Romain
- Implementation on the ComputeTortuosity module (only Distance Metric)
- Started debugging
- Future
- More debugging! ---> hopefully testing soon
- #tube command
- Implementation of the other 2 methods : ICM SOAM
Danielle
Sliding organ registration
- Fixed "duh" problem from Thursday - extracting tangential and normal components before computing regularization
- Regularization smoothing in normal direction completed
- Added intensity distance function (mean squares)
Translated sphere - motion field following registration WITHOUT regularization:
Translated sphere - motion field following registration WITH regularization:
XCAT phantom
- Got XCAT running on my machine (thanks Andinet!)
- Learned XCAT parameters
- Wrote isbiValidation.py:
- --basic (XCAT mode 0 - phantom mode): with respiratory period 5s and cardiac period 1s, generates two images at 0s and 2.5 s, saves as .mhd
- --gold (XCAT mode 2 - spherical lesion generation mode): creates gold standard motion field, and saves as .txt (index, vector in 6 columns). Then run itkImageToImageDiffusiveDeformableRegistrationGenerateGoldStandardTest to create itk::Image<VectorType, Dimension> .mhd for visualization
- XCAT phantom gives gold standard vectors for surface borders (XCAT mode 4), but no internal vectors! So...
- for index in range - generate spherical lesion at index, use nurbs_save = 1 to save lesion.nrb, then calculate centroid of lesion control points
- Question: are there discontinuities / sliding motion. Looks like it =) See circled region:
- Question: does the motion field make sense? Yes:
Fixed image (look at origin of vectors):
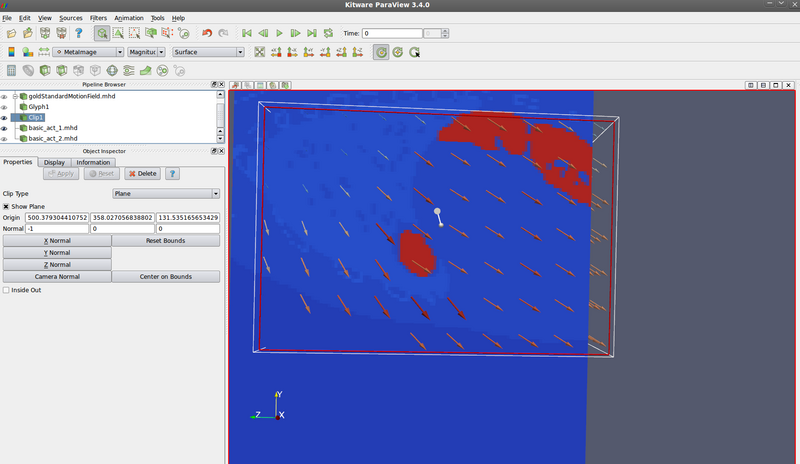
Moving image (look at destination of vectors):
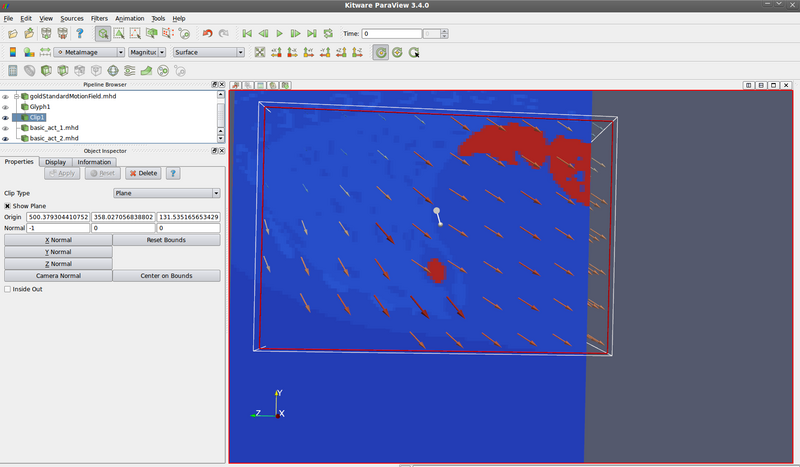
- --segmentation (XCAT mode 5 - save anatomical variation mode, or run XCAT mode 0 with save_nurbs = 1) to save surfaces (.nrb text file). Then create new .txt file for the surface you want. Then run itkImageToImageDiffusiveDeformableRegistrationGenerateNormalsTest to create itk::Image<PixelType, Dimension> of control points
- Unfortunately: (1) control points need interpolation, can't find NURBS support in VTK; (2) XCAT has MANY types of surfaces for one organ - ex all intracardiac surfaces, etc, and it would be a lot of work to sort through them; (3) Perfect segmentation not great for registration method evaluation - removes segmentation error component of total error.
- Ex: right lung
- --segmentation (XCAT mode 5 - save anatomical variation mode, or run XCAT mode 0 with save_nurbs = 1) to save surfaces (.nrb text file). Then create new .txt file for the surface you want. Then run itkImageToImageDiffusiveDeformableRegistrationGenerateNormalsTest to create itk::Image<PixelType, Dimension> of control points
Talking points
- Underlying XCAT deformation fields too perfect? They are likely interpolating the border vectors.
Future
- Boundary conditions
- Compute weighting term = distance to organ border
- Extract organ border normals from input data
- Run registration + compare to gold standard for validation #s