Particle simulations: Difference between revisions
No edit summary |
|||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
=Introduction= | =Introduction= | ||
Particle simulations | Particle simulations consist of point data, rather than cell or element data. An example may be electrons in a field, or water molecules in a fluid. Particle datasets can be combined with other datasets to show particle motion in a solid or liquid. | ||
This tutorial uses a dataset named mediumVector.vtu. I will try to get it on the Kitware web site. | This tutorial uses a dataset named mediumVector.vtu. I will try to get it on the Kitware web site. If you want to make it yourself, it is just a vtk dataset, using the format listed at the end of this tutorial. I then read it into ParaView, and used the reflect filter in the X, then Y, then Z direction. I believe I then did the same reflections again. Finally, this file was written out to disk as a .vtu file. | ||
=Displaying particles= | =Displaying particles= | ||
* Open ParaView, and read in your dataset. | * Open ParaView, and read in your dataset. | ||
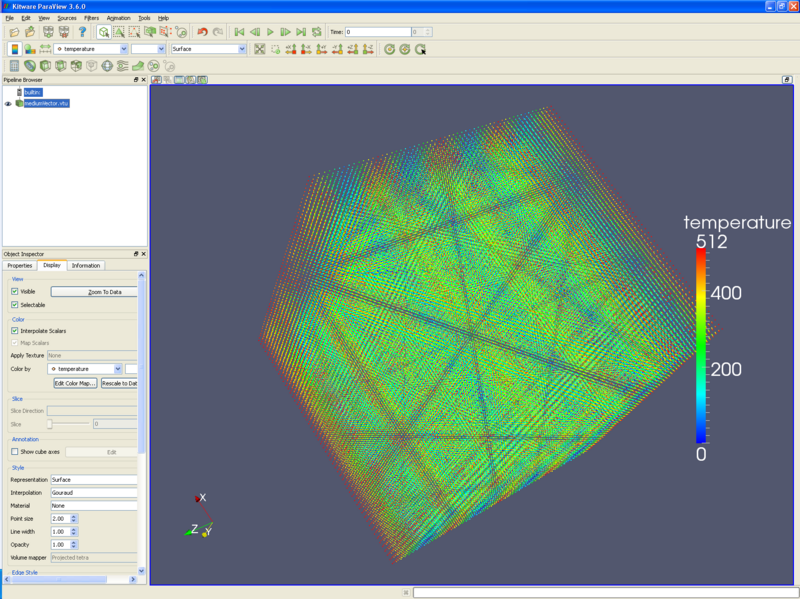
* If you are running ParaView 3.6.0 or 3.4.0, you will want to reset the point sizes. Display tab/ Style/ Point size 2 (or 1, depending on your preference). The following picture holds about 250,000 points. | * If you are running ParaView 3.6.0 or 3.4.0, you will want to reset the point sizes. '''Display tab/ Style/ Point size''' 2 (or 1, depending on your preference). The following picture holds about 250,000 points. | ||
[[Image:particles_1.png|800px]] | [[Image:particles_1.png|800px]] | ||
* You may want to replace these particles with spheres or arrows, using the glyph filter. The glyph filter will depopulate your dataset to a reasonable number of items, set by the | |||
=Glyphing particles= | |||
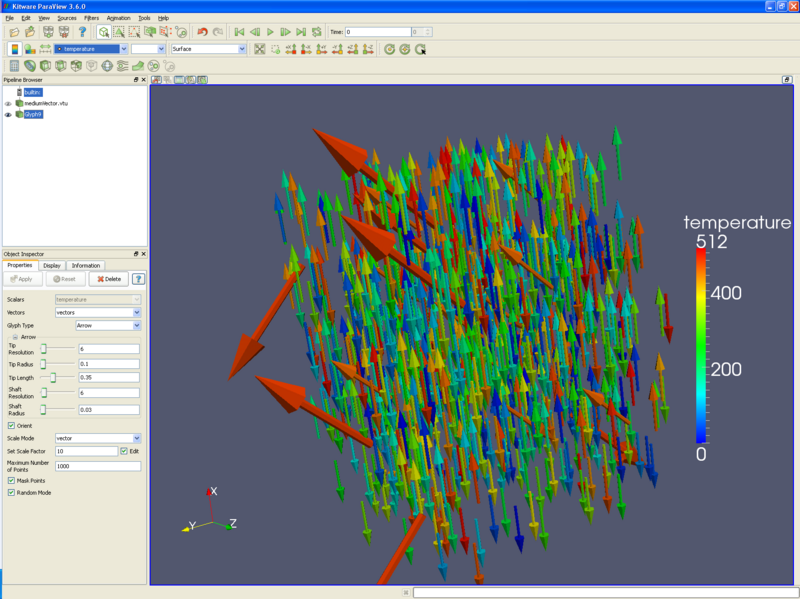
* You may want to replace these particles with spheres or arrows, using the glyph filter. The glyph filter will depopulate your dataset to a reasonable number of items, and this reasonable number is set by the '''Maximum number of points''' input box. '''Random mode''' often creates better looking output, but is not useful if you animate through numerous time steps. The following picture has arrows for particles, using vectors for direction and length, and colored by temperature. | |||
[[Image:particles_2.png|800px]] | [[Image:particles_2.png|800px]] | ||
* Another filter that is useful is the threshold filter. Using the threshold filter, a user can select only those particles that are of interest. For instance, the following picture only displays particles that have a temperature of 430 to 511 degrees. Using the clip filter, the upper half of the picture has arrow glyphs, and the lower half has the particles themselves - all colored by temperature. | |||
=Thresholding particles= | |||
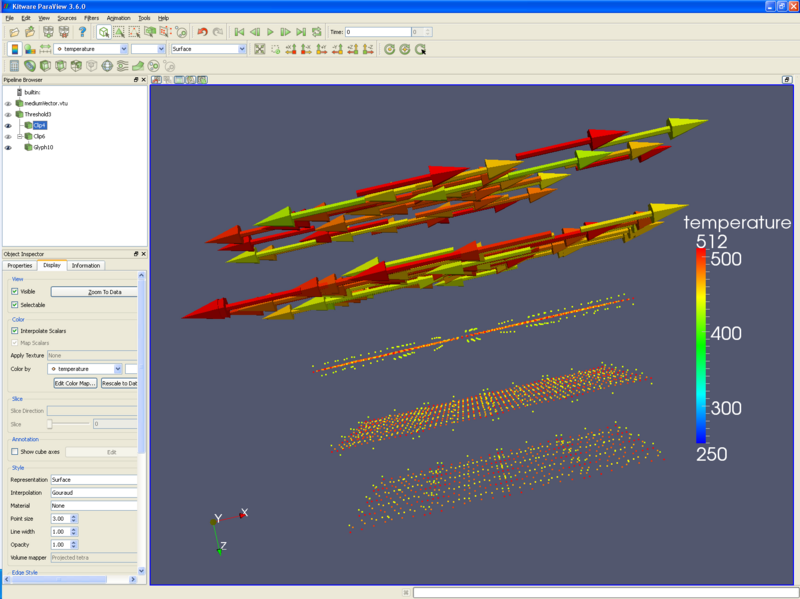
* Another filter that is useful is the '''threshold filter'''. Using the '''threshold filter''', a user can select only those particles that are of interest. For instance, the following picture only displays particles that have a temperature of 430 to 511 degrees. Using the '''clip filter''', the upper half of the picture has arrow glyphs, and the lower half has the particles themselves - all colored by temperature. | |||
[[Image:particles_3.png|800px]] | [[Image:particles_3.png|800px]] | ||
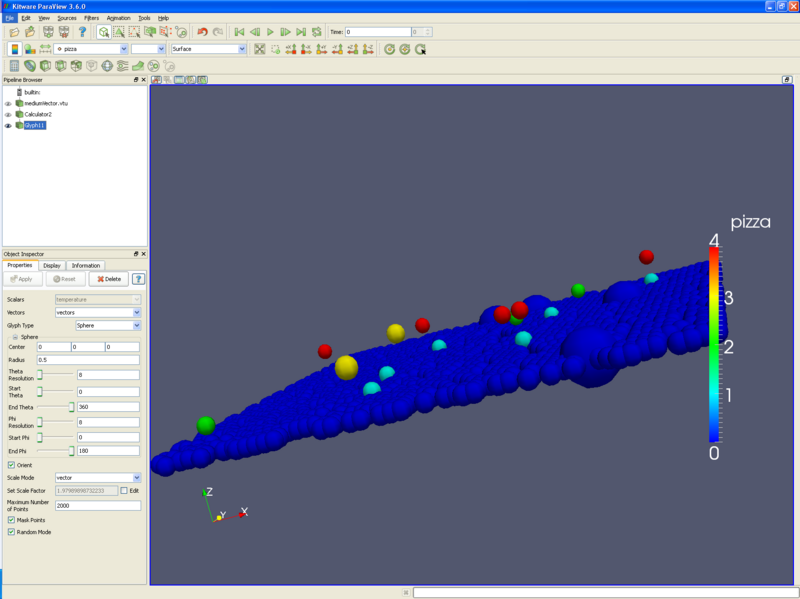
*Finally, you can make a scatter plot through the calculator. For instance, if we want to preserve the X and Y coordinates of our points, but use number of pizzas for the Z component, we would do the following: | |||
**Open the calculator filter. | =Scatter Plot= | ||
**Check the Coordinate Results check box. | *Finally, you can make a scatter plot through the '''calculator filter'''. For instance, if we want to preserve the X and Y coordinates of our points, but use number of pizzas for the Z component, we would do the following: | ||
**Input the formula coordsX*iHat+coordsY*jHat+pizza*kHat. This creates a vector, as follows: (X,Y,pizzas) | **Open the '''calculator filter'''. | ||
**Check the '''Coordinate Results''' check box. | |||
**Input the formula '''coordsX*iHat+coordsY*jHat+pizza*kHat'''. This creates a vector, as follows: (X,Y,pizzas). (Think of the iHat as "This is an X", the * as tying the coordinates and variables together, and the + sign as the comma between X,Y, and Z.) | |||
[[Image:particles_4.png|800px]] | [[Image:particles_4.png|800px]] | ||
| Line 31: | Line 41: | ||
=Example data file= | =Example data file= | ||
This data is written | This data is written to a .vtk file. Note that I have deleted a lot of the data, which will need to be recreated. Also note that I added some random variable data - for instance, some locations have 1 or 2 pizzas. This actually creates a strange dataset - with many points per cell, but it worked for display purposes. | ||
<source lang="text"> | |||
# vtk DataFile Version 2.0 | # vtk DataFile Version 2.0 | ||
Unstructured Grid Example | Unstructured Grid Example | ||
| Line 53: | Line 64: | ||
... | ... | ||
POINT_DATA 512 | POINT_DATA 512 | ||
SCALARS | SCALARS temperature float 1 | ||
LOOKUP_TABLE default | LOOKUP_TABLE default | ||
000.0 001.0 002.0 003.0 004.0 005.0 006.0 007.0 008.0 009.0 | 000.0 001.0 002.0 003.0 004.0 005.0 006.0 007.0 008.0 009.0 | ||
| Line 66: | Line 77: | ||
1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 | 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 | ||
1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 | 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 | ||
... | |||
</source> | |||
| Line 72: | Line 84: | ||
=Acknowledgements= | =Acknowledgements= | ||
Sandia is a | Sandia National Laboratories is a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA-0003525. | ||
Latest revision as of 20:37, 22 June 2017
Introduction
Particle simulations consist of point data, rather than cell or element data. An example may be electrons in a field, or water molecules in a fluid. Particle datasets can be combined with other datasets to show particle motion in a solid or liquid.
This tutorial uses a dataset named mediumVector.vtu. I will try to get it on the Kitware web site. If you want to make it yourself, it is just a vtk dataset, using the format listed at the end of this tutorial. I then read it into ParaView, and used the reflect filter in the X, then Y, then Z direction. I believe I then did the same reflections again. Finally, this file was written out to disk as a .vtu file.
Displaying particles
- Open ParaView, and read in your dataset.
- If you are running ParaView 3.6.0 or 3.4.0, you will want to reset the point sizes. Display tab/ Style/ Point size 2 (or 1, depending on your preference). The following picture holds about 250,000 points.
Glyphing particles
- You may want to replace these particles with spheres or arrows, using the glyph filter. The glyph filter will depopulate your dataset to a reasonable number of items, and this reasonable number is set by the Maximum number of points input box. Random mode often creates better looking output, but is not useful if you animate through numerous time steps. The following picture has arrows for particles, using vectors for direction and length, and colored by temperature.
Thresholding particles
- Another filter that is useful is the threshold filter. Using the threshold filter, a user can select only those particles that are of interest. For instance, the following picture only displays particles that have a temperature of 430 to 511 degrees. Using the clip filter, the upper half of the picture has arrow glyphs, and the lower half has the particles themselves - all colored by temperature.
Scatter Plot
- Finally, you can make a scatter plot through the calculator filter. For instance, if we want to preserve the X and Y coordinates of our points, but use number of pizzas for the Z component, we would do the following:
- Open the calculator filter.
- Check the Coordinate Results check box.
- Input the formula coordsX*iHat+coordsY*jHat+pizza*kHat. This creates a vector, as follows: (X,Y,pizzas). (Think of the iHat as "This is an X", the * as tying the coordinates and variables together, and the + sign as the comma between X,Y, and Z.)
Example data file
This data is written to a .vtk file. Note that I have deleted a lot of the data, which will need to be recreated. Also note that I added some random variable data - for instance, some locations have 1 or 2 pizzas. This actually creates a strange dataset - with many points per cell, but it worked for display purposes.
<source lang="text">
- vtk DataFile Version 2.0
Unstructured Grid Example ASCII
DATASET UNSTRUCTURED_GRID POINTS 512 float 0 0 0 1 0 0 2 0 0 3 0 0 4 0 0 5 0 0 6 0 0 7 0 0 0 1 0 1 1 0 2 1 0 3 1 0 4 1 0 5 1 0 6 1 0 7 1 0 ... 0 0 1 1 0 1 2 0 1 3 0 1 4 0 1 5 0 1 6 0 1 7 0 1 0 1 1 1 1 1 2 1 1 3 1 1 4 1 1 5 1 1 6 1 1 7 1 1 ... 0 0 2 1 0 2 2 0 2 3 0 2 4 0 2 5 0 2 6 0 2 7 0 2 0 1 2 1 1 2 2 1 2 3 1 2 4 1 2 5 1 2 6 1 2 7 1 2 ... CELL_TYPES 52 1 1 ... POINT_DATA 512 SCALARS temperature float 1 LOOKUP_TABLE default 000.0 001.0 002.0 003.0 004.0 005.0 006.0 007.0 008.0 009.0 010.0 011.0 012.0 013.0 014.0 015.0 016.0 017.0 018.0 019.0 ... SCALARS pizza float 1 LOOKUP_TABLE default 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 000.0 ... VECTORS vectors float 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 1 0 0 ... </source>
Acknowledgements
Sandia National Laboratories is a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA-0003525.